Collaborators
Internal MembersLuca Fiaschi, Dr. Ferran Diego, Dr. Fred A. Hamprecht
External CollaboratorsDr. Juergen Reymann (Bioquant - Heidelberg, Germany), Dr. Ana Martin-Villalba (DKFZ - Heidelberg, Germany), Dr. Andreas Draguhn (IZN -Heidelberg, Germany), Dr. Marta Zlatic (HHMI - Janelia Farm, USA), Dr. Daniel W. Gerlich (ETH - Zurich, Switzerland).
Research Focus
New microscopic imaging techniques pose both grand challenges and new opportunities for image processing and computer vision. At the same time, the analysis of high-throughput data generated in biological research requires novel mathematical and algorithmic methods in order to scale to massive image databases. Working at the interface of image processing, statistics and machine learning, we develop, in close collaboration with biologists, tools to analyze high-throughput microscopy images. The major requirements in the field of high-throughput image analysis are described by the following research lines:[ Quality Control ], [ Cell Detection ], [ Structured Learning ].
Weakly Supervised Learning for Image Quality Control
In large scale bio medical experiments, the quality of the resulting data is crucial for any meaningful analysis. In fact, defect images jeopardize downstream analysis, yielding misleading results or even false conclusions. Image defects can occur during sample preparation, such as debris contamination or during image acquisition, such as out-of-focus images. In several applications, it is important to distinguish between regional defects and global defects: for example, an out-of-focus image can be easily corrected during acquisition by refocusing the microscope.
 |
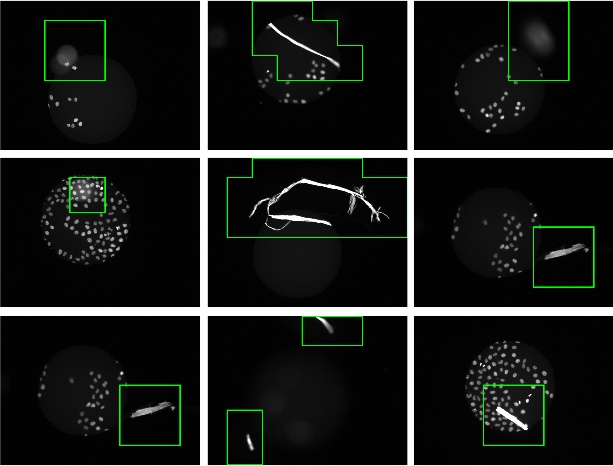 |
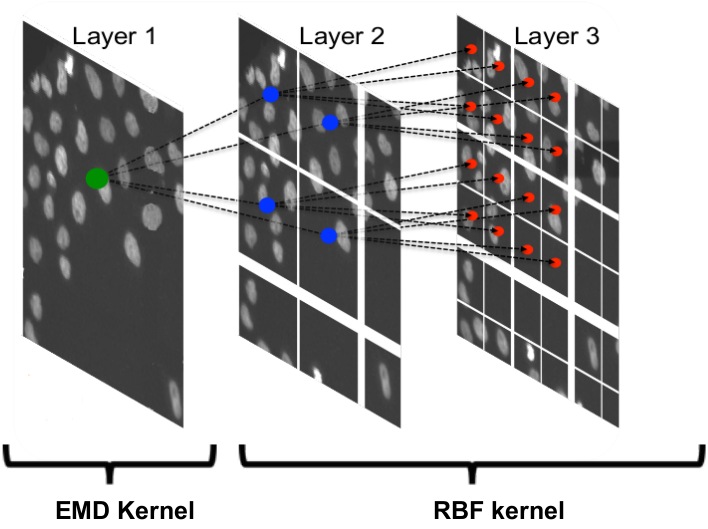
Our approach casts the problem of defect image classification as an outlier detection task (i.e. defect images as outliers) based on the one-class SVM algorithm. To exploit the characteristics of global and regional defects we handle the two classes in different hierarchical stages. Stage one operates on the full image level and aims at filtering out globally defect images such as out-of-focus. Stages two and three work on patch level and form a coarse-to-fine procedure for detecting regional defects. The overall framework resembles a cascade of one-class SVM classifiers.
- X. Lou, L. Fiaschi, U. Koethe, F. A. Hamprecht, Quality Classification of Microscopic Imagery with Weakly Supervised Learning, MICCAI-MLMI. Proceedings, (2012) best paper award winner [Technical Report]
Mitotic Cell Detection in Histopathological Images
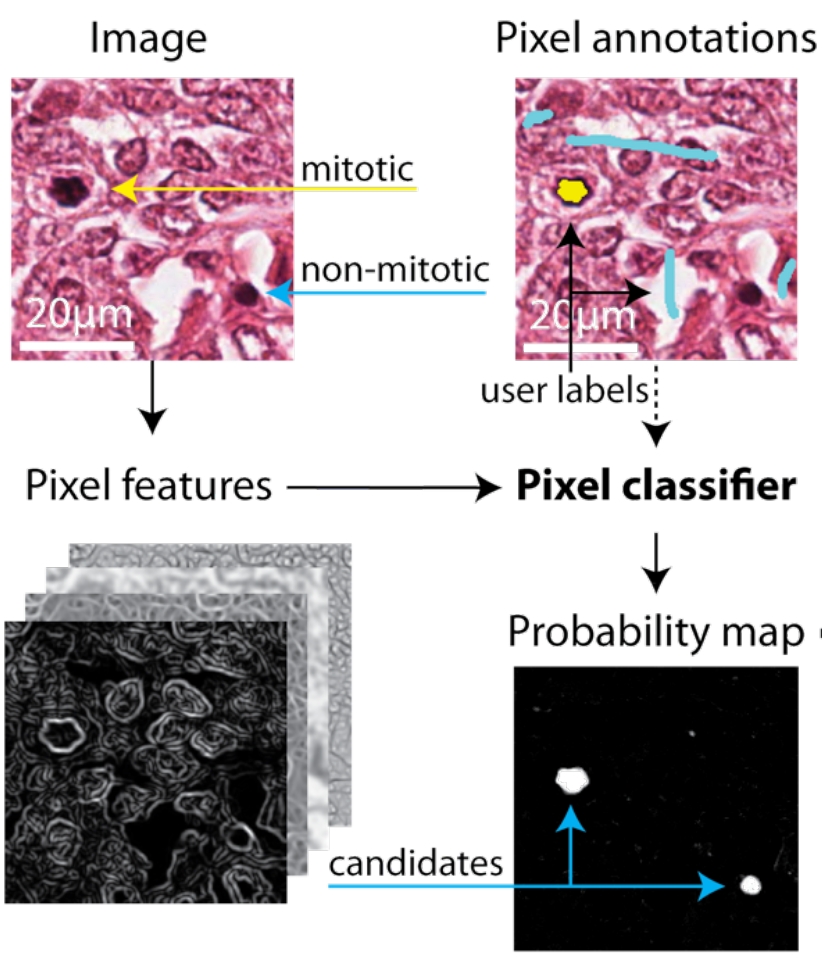
The mitotic cell index is currently the best predictor for long term prognosis of breast carcinomas. Despite recent advancements in automatic imaging of biopsy slides, the manual counting of the mitotic cells is still today the standard procedure in clinical practice. In order to make pathologist everyday work easier, we provide them with a learning based workflow for semi-automated mitosis detection.
Our approach is based on a hierarchy of classifiers. In the first stage a pixel-wise classifier is trained to segment candidate cell regions from an initial set of annotated training images. Cells candidates are then classified into mitotic or non-mitotic cells using global features of the foreground region, such as shape-context, shape moments, and statistical texture features. Our workflow banks on two open-source biomedical image analysis software: ilastik and CellCognition, which provide an user user friendly interface to state of the art learning algorithms.
References
- C. Sommer, L. Fiaschi, F. A. Hamprecht, D. Gerlich,
Learning-based Mitotic Cell Detection in Histopathological Images, ICPR, Proceedings, (2012) [Technical Report] - C. Sommer, C. Straehle, U. Koethe, F. A. Hamprecht
ilastik: Interactive Learning and Segmentation Toolkit ,ISBI, Proceedings, (2011), 230-233 [Technical Report]
Structured Dictionary Learning for Identification of Neuronal Assemblies in Calcium Imaging
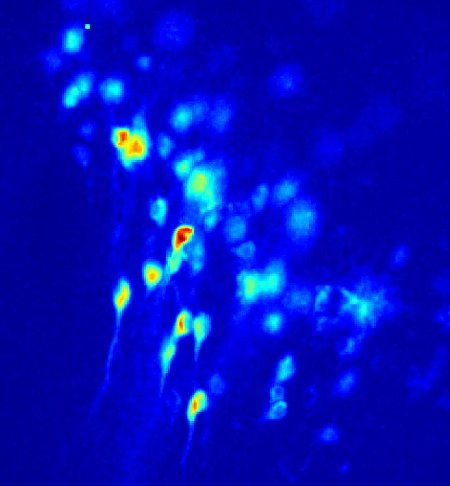
Fluorescent calcium imaging allows monitoring the spatio-temporal co-activation of hundreds of neurons in rapidly oscillating three-dimensional networks. This makes it an essential tool for studying complex patterns of distributed activity in neuronal networks such as the processes of decision making or consciousness. The ultimate goal of our research is the identification and characterization of stable neuron activation patterns, termed neural assemblies. This entails the detection of cell centroids and the identification of calcium transients (events) that reappeared during different activity periods.
Since these events are sparsely distributed in time and space, we model the problem as the construction of a structured sparse dictionary. We are currently developing efficient algorithms to solve the large optimization problem which arises from sparse structured dictionary learning on an high throughput data set. Our method is conveniently implemented in an open source toolbox to support biologists' work.


