Abstract
This 3D+t dataset shows an excerpt of a developing fruit fly (Drosophila melanogaster) embryo over 100 time steps during gastrulation. The movie has been recorded using a light sheet microscope. We provide dense manual annotations for cell lineages for all ~800 cells per frame.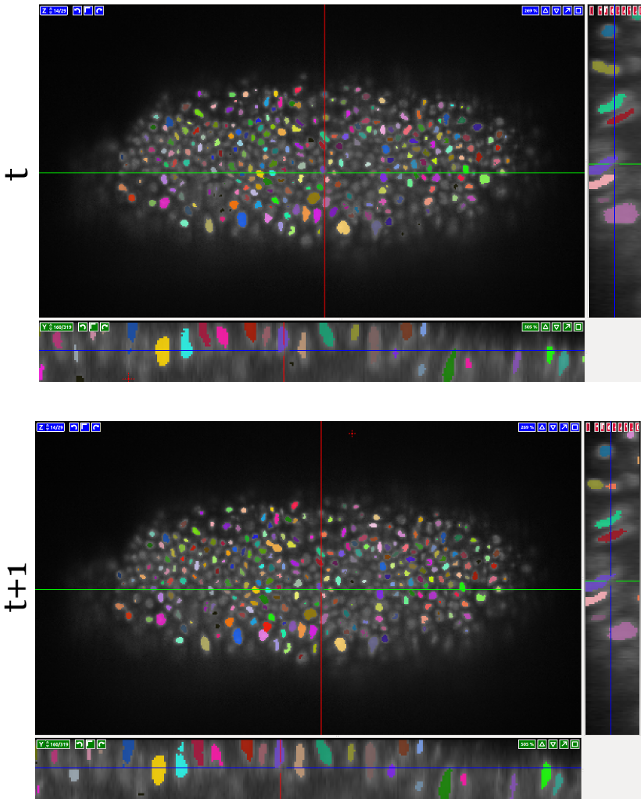
The movie of this developing Drosophila melanogaster embryo has been recorded by the Hufnagel group, EMBL Heidelberg, Germany, with the light sheet microscope described in (Krzic et al., 2012).
The challenging task is to automatically segment and track all cells over all time steps to reconstruct full cell lineages.
To acquire manual cell tracking annotations, we first segmented this dataset using ilastik and refined the result with a seeded watershed. On average, this yielded ~800 cells per frame, which we then tracked manually over all 100 time steps using the Manual Tracking workflow in ilastik. We here provide the raw data, our segmentation and our manual annotations for benchmarking purposes.
This data may be used for research purposes only. When you use this dataset in any publication, please cite this article:
M. Schiegg*, P. Hanslovsky*, C. Haubold, U. Koethe, L. Hufnagel, F. A. Hamprecht. Graphical Model for Joint Segmentation and Tracking of Multiple Dividing Cells. Bioinformatics, in press, 2015. [* contributed equally]
|
@Article{schiegg_15_graphical, |
Technical details:
- Organism: Drosophila melanogaster embryo, gastrulation stage
- Fluorescent protein: H2Av-mCherry
- Microscope: Multiview selective-plane illumination microscope (MuVi-SPIM) (Krzic et al. 2012)
- Volume dimensions: 730 × 320 × 30 voxels
- Voxel size: 0.52μm × 0.52μm × 0.52μm (3D isotropic)
- Temporal resolution: 1 frame per 30 seconds
- Number of frames: 100
Note: Making dense ground-truth in 3D+t is a very challenging task. We share here our manual annotations and would appreciate any contributions in order to further improve the gold standard for this challenging dataset. Please contact the authors if you want to contribute.
Content
We are providing the following contents. Our recommended file type is HDF5 (available for Matlab, Python, C++, etc. or with a GUI), but for convenience, we provide all images as multipage tiff sequences, too.
- ./raw_data/: Raw Data: uint16, 100 frames, 730 × 320 × 30 voxels
- ./segmentation/: Our binary segmentation files, uint8
- ./object_identities/: The label images contain for each voxel the assigned object identity (referred to as oid), uint32
- ./manual_annotations/manual_annotations.h5: Our manual cell tracking annotations are provided in this hdf5 file in the following format. Use e.g. hdfview to open this dataset.
Note that the lineage tree of one cell may consist of multiple tracks which are connected through divisions.
- table: contains the following columns
- object_id: a unique running identifier
- timestep: the frame number
- labelimage_oid: the object identifier, unique for each object in each frame, as in ./object_identities/.
- track_id1: unique track identifier (started by appearance or cell division; terminated by disappearance or cell division)
- Count: The number of voxels assigned to this cell (its size).
- Coord<Minimum>_N: The lower left corner of the 3D bounding box around the object (N=1,2,3 for dimensions x,y,z)
- RegionCenter_N: The center of the segmented cell. (N=1,2,3 for dimensions x,y,z)
- Coord<Maximum>_N: The upper right corner of the 3D bounding box around the object (N=1,2,3 for dimensions x,y,z)
- divisions: contains the following columns
- timestep: the frame number
- parent_oid: the object identifier of the parent cell in the given timestep
- track_id: the track identifier of the parent cell in the given timestep
- child1_oid: the object identifier of one daughter cell in the given timestep+1
- child1_track1_id: the track identifier of this daughter cell in the given timestep+1
- child2_oid: as above
- child2_track1_id: as above
- images/object_id/raw: the raw data in the 3D bounding box of this object
- images/object_id/labeling: the segmentation mask in the 3D bounding box of this object (binary image)
- ./manual_annotations/convenience_files/: For convenience, we additionally export the cell tracking annotations provided in manual_annotations.h5to the following formats:
- ./CSV/: The table and divisions datasets from./manual_annotations/manual_annotations.h5 exported as csv tables. See above for descriptions.
- ./track_identities/: Sequences of 3D images in which each voxel contains its track identifier. For divisions, refer to the divisions tables described above.
- ./H5-Events-Sequence/: One hdf5 file per timestep. For further details on this format, we refer to the ilastik user documentation.
- ./drosophila_manualTracking-ilastik.ilp: ilastik project file for the manual tracking workflow. All files above have been generated from ilastik using this project file.


