Marc Kirchner, Bernhard Y. Renard, Michael Hanselmann, Xinghua Lou, Anna Kreshuk, Bernhard Kausler, Ullrich Köthe, Fred A. Hamprecht,
in close collaboration with- Steen & Steen Lab, Children's Hospital Boston
- Systems Biology, Harvard Medical School.
- Mayer Lab, Zentrum für Molekulare Biologie der Universität Heidelberg
- Heeren Group, Institute for Atomic and Molecular Physics, Amsterdam, The Netherlands
- Mass Spectrometry Group, Molecular Structure Analysis, DKFZ
- Helmholtz Zentrum München
- Klinikum rechts der Isar, München
- Hanselmann M, Köthe U, Kirchner M, Renard BY, Amstalden ER, Glunde K, Heeren RMA, Hamprecht FA (2009). Toward Digital Staining using Imaging Mass Spectrometry and Random Forests. Journal of Proteome Research, 8(7), 3558-3567
- Renard BY*, Kirchner M*, Steen H, Steen JAJ, Hamprecht FA (2008). NITPICK: Peak Identification for Mass Spectrometry Data, BMC Bioinformatics 2008, 9:355. [full text]
- Steen JAJ, Steen H, Georgi A, Parker K, Springer M, Kirchner M, Hamprecht FA, Kirschner MW (2008). Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: A quantitative proteomic analysis, Proceedings of the National Academy of Sciences (PNAS), 105(16), pp. 6069-6074 . [pdf]
- Hanselmann M, Kirchner M*, Renard BY*, Amstalden ER, Glunde K, Heeren RMA, Hamprecht FA (2008). Concise Representation of Mass Spectrometry Images, Analytical Chemistry 80(24):9649-9658.
- Koenig T, Menze BH, Kirchner M*, Monigatti F*, Parker KC*, Patterson T, Steen JAJ, Hamprecht FA, Steen H (2008). Robust Prediction of the MASCOT Score for an Improved Quality Assessment in Mass Spectrometric Proteomics, Submitted.
- Kirchner M, Saussen B, Steen H, Steen JAJ, Hamprecht FA. (2007) "amsrpm: Robust Point Matching for Retention Time Alignment of LC/MS Data with R", Journal of Statistical Software, Special Issue on Spectroscopy and Chemometrics in R, 18(4) [pdf]
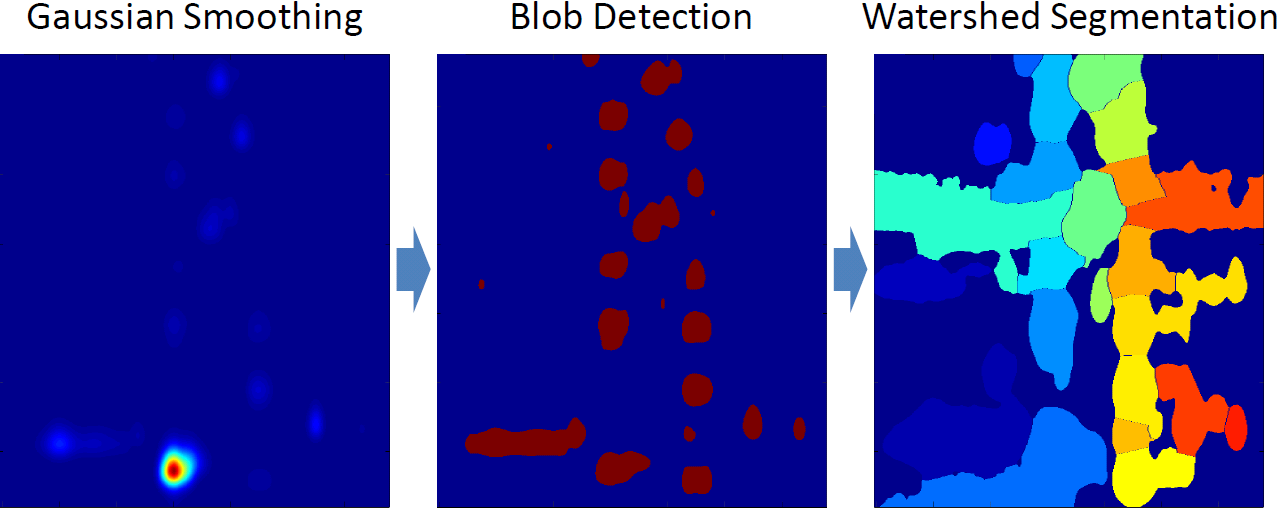
- Hanselmann M, Köthe U, Renard BY, Kirchner M, Heeren RMA, Hamprecht FA (2009). Multivariate Watershed Segmentation of Compositional Data. Proc. of the 15th Int. Conf. on Discrete Geometry for Computer Imagery (DGCI) (accepted for publication).
- Kirchner M*, Renard BY*, Koethe U, Steen JAJ, Steen H, Hamprecht, FA. (2009). Automatic Computational Protein Co-Regulation Screening for Quantitative Mass Spectrometry Experiments. Conference of the American Society for Mass Spectrometry (ASMS) 2009
- Kreshuk A, Kirchner M, Renard BY, Winter D, Steen H, Steen JAJ, Lehmann WD, Hamprecht FA. (2009) . Automatic Quantification of 16/18O-Labeled LC/MS Data. Conference of the American Society for Mass Spectrometry (ASMS) 2009
- Voss BM, Renard BY, Kreshuk A, Hanselmann M, Koethe U, Steen H, Steen JAJ, Kirchner M, Hamprecht FA. (2009). Simultaneous Multiple Alignment for LC/MS Peak Lists. Conference of the American Society for Mass Spectrometry (ASMS) 2009
- Lou X, Kirchner M*, Renard BY*, Koethe U, Graf C, Steen JAJ, Steen H, Mayer MP, Hamprecht FA. (2009). HeXicon++: Automating HDX Data Analysis. Conference of the American Society for Mass Spectrometry (ASMS) 2009
- Renard BY, Monigatti F, Kirchner M, Ivanov AR, Rappsilber J, Steen JAJ, Hamprecht FA, Steen H. (2009). Computational MS/MS Spectra Preprocessing - a Free Lunch. Conference of the American Society for Mass Spectrometry (ASMS) 2009
- Hanselmann M, Koethe U, Kirchner M*, Renard BY*, Amstalden ER, Glunde K, Heeren RMA, Hamprecht FA. (2009). Automated Classification and Grading of Tumors in Mass Spectrometric Images using postprocessed Random Forests. Conference of the American Society for Mass Spectrometry (ASMS) 2009
- Lou X, Kirchner M*, Renard BY*, Graf C, Steen JAJ, Steen H, Mayer MP, Hamprecht FA. (2008). HeXion: Automatic Information Extraction for HX-MS Data, HX interest group meeting, Conference of the American Society for Mass Spectrometry (ASMS) 2008.
- Kirchner M*, Renard BY*, Monigatti F, Hamprecht FA, Steen JAJ, Steen H. (2008). Optimal Significane Determination in iTRAQ experiments, Conference of the American Society for Mass Spectrometry (ASMS) 2008.
- Renard BY*, Kirchner M*, Steen JAJ, Steen H, Hamprecht FA. (2008). STRAP2: Reliable, Hierarchical Feature Extraction from Multicomponent Mass Spectra, Conference of the American Society for Mass Spectrometry (ASMS) 2008.
- Hanselmann M, Kirchner M, Renard BY, Kharchenko A, Klerk L, Koethe U, Heeren RMA, Hamprecht FA. (2008) Concise representation of MS Images by Probabilistic Latent Semantic Analysis, Conference of the American Society for Mass Spectrometry (ASMS) 2008.
- Boppel S, Renard BY, Kirchner M, Steen H, Koethe U, Hamprecht FA. (2008). Sparse Profile Reconstruction for LC/MS Feature Extraction, Conference of the American Society for Mass Spectrometry (ASMS) 2008.
- Lou X, Kirchner M*, Renard BY*, Koethe U, Steen H, Mayer MA, Hamprecht FA. (244008). Fully Automated HX-MS Data Analysis with Complete Deuteration Distribution Estimation, Conference of the American Society for Mass Spectrometry (ASMS) 2008.
- Renard BY, Kirchner M, Steen JAJ, Steen H, Hamprecht FA (2007). Extended Fractional Averagine for Improved Peak Identification in Mass Spectrometry Data, European Conference on Computational Biology (ISMB/ECCB) 2007
- Kirchner M, Renard BY, Goerlitz L, Steen H, Steen JAJ, Hamprecht FA (2007). A sparsity-promoting non-negative regression approach for deconvolution, deisotoping, and quantification of mixture mass spectra, American Society for Mass Spectrometry, Annual Conference, ASMS 2007
- Renard BY, Kirchner M, Steen JAJ, Steen H, Hamprecht FA (2007). Fractional Averagine: Improved Modeling Of Isotope Distributions For Peak Picking. American Society for Mass Spectrometry, Annual Conference, ASMS 2007.
- Koenig T, Menze BH, Kirchner M, Steen H, Steen JAJ, Monigatti F, Parker KC, Patterson T, Hamprecht FA (2007). A Quality Score for Product Ion Spectra in Proteomics: How to Teach an Expert System with Minimal Manual Input, American Society for Mass Spectrometry, Annual Conference, ASMS 2007
- Steen H, Steen JAJ, Parker K, Georgi A, Ross P, Kirchner M, Hamprecht FA, Pappin D, Kirschner MW. (2006) Deciphering the Cell Cycle Through Quantitative Proteomics, 17th International Mass Spectrometry Conference (IMSC) 2006
- Bonowski F, Kirchner M, Steen H, Steen JAJ, Hamprecht FA. (2006) "ProteoPeak" - a modular framework for intelligent peak picking in biopolymer samples using adaptive templates, 17th International Mass Spectrometry Conference (IMSC) 2006
- Saussen B, Kirchner M, Steen JAJ, Steen H, Hamprecht FA. (2006) The rpm package: aligning LC/MS mass spectra with R, useR!2006.
- Waldon ZO, Kirchner M, Bonowski F, Hamprecht FA, Steen H, Jebanathirajah J. (2006) Discriminatrix: A System for Discovering Microbial Biomarkers, American Society for Mass Spectrometry, Annual Conference, ASMS 2006.
Overview
The tasks in MS data processing have roots in many fields including signal and image processing, statistical modeling, decision theory, statistical learning, and pattern recognition. Within this rapidly moving field we focus on providing robust automated methods for quantitative analysis.
Feature extraction
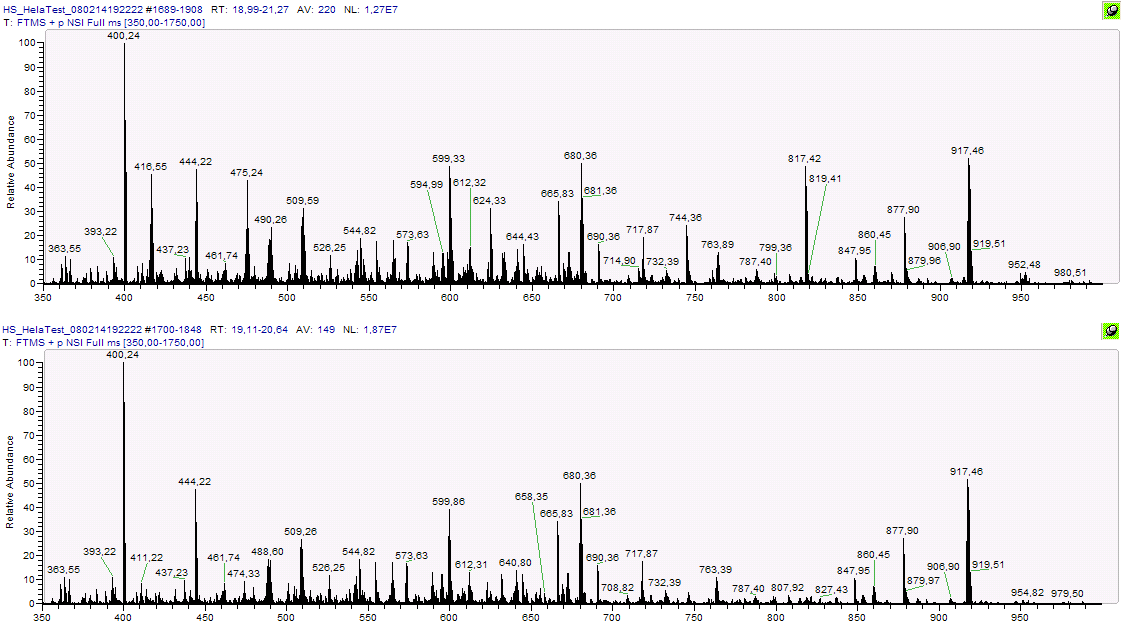
Most research efforts on the analysis of mass spectrometric data share a common approach based on post-acquisition signal filtering - followed by peak picking, alignment and statistical modeling. Sound methods exist for each of these steps, but mere pipelining neither seems to be able to cope with the complexity present in LC/MSn data nor does it provide access to more than a fraction of the overwhelming amount of information buried within the high dimensional data, thus calling for new methods.
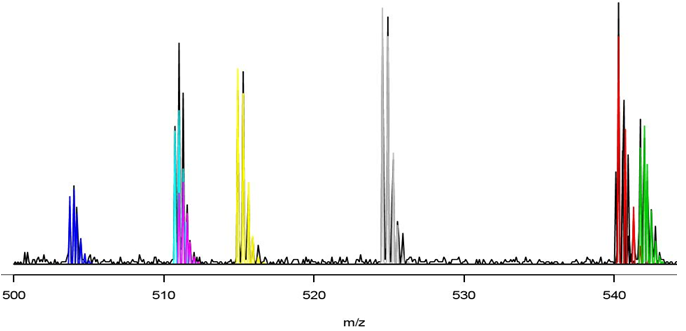
Biomarker detection
From a statistical point of view, finding biomarkers can be described as finding those peaks in the mass spectrum that maximize discrimination between species. Biomarkers need to be as sensitive as possible to avoid false negatives and also as specific as possible to avoid false positives. The problem is nontrivial since biomarker condidates need to be found among tens of thousands of m/z values with only a few hundred samples available.