We aim at novel methodology for solving large-scale multi-matching problems in an unsupervised fashion.
Such problems emerge in applications in biology in the context of organisms that are stereotypical at the level of individual cells.
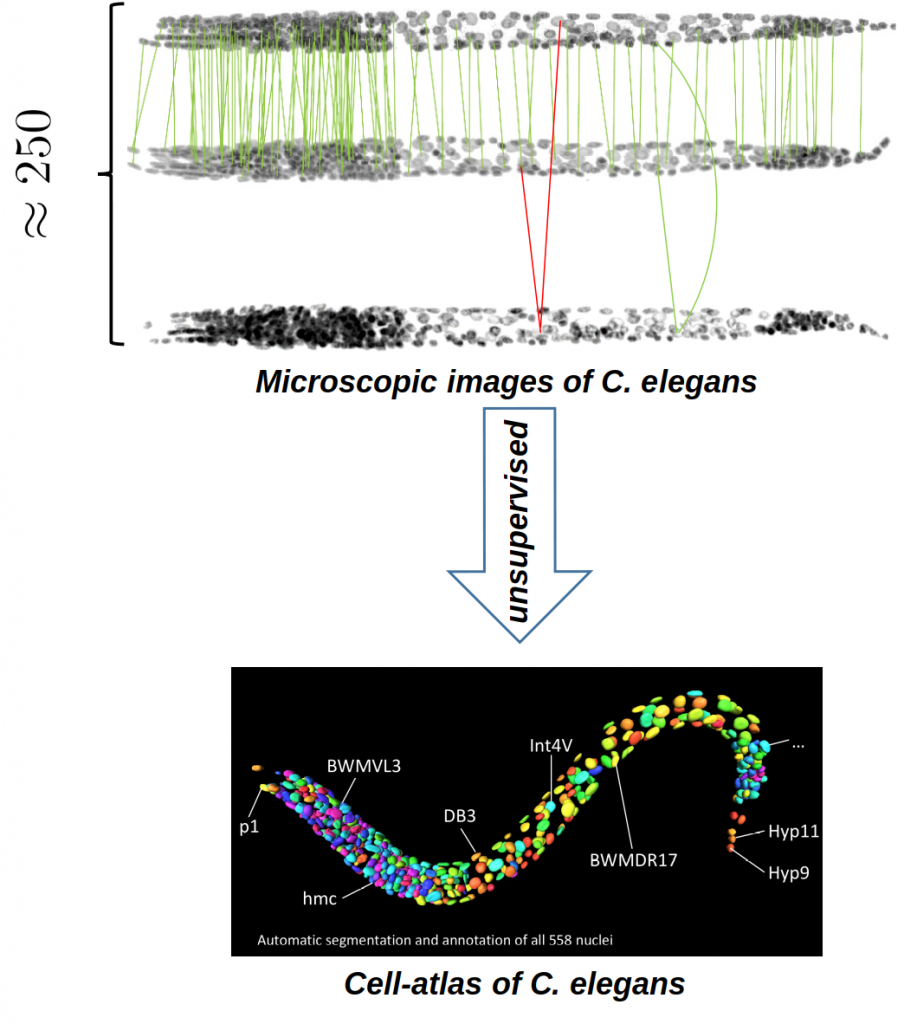
In particular, our project addresses the popular model organism Caenorhabditis elegans , a roundworm that has 558 cells at its first larval stage. The project is supported by DFG Projects 98181230 and 53943535 and is run together with Kainmüller Lab in Max Delbrück Center for Molecular Medicine, Berlin.
Steps done so far
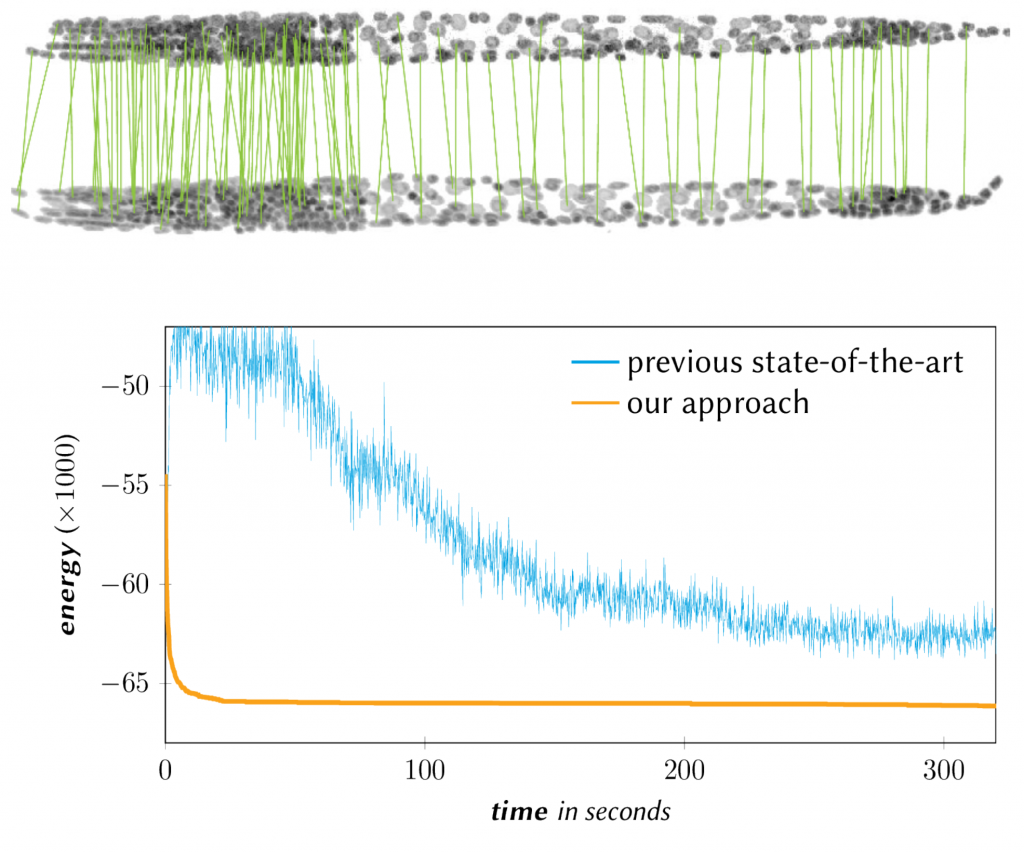
Fast scalable graph matching algorithm
L.Hutschenreiter, S. Haller, L. Feineis, C. Rother, D. Kainmueller, B. Savchynskyy Fusion Moves for Graph Matching ICCV 2021 [pdf] [project page] [slides] [poster]
S. Haller, L. Feineis, L.Hutschenreiter, C. Rother, D. Kainmueller, P. Swoboda, B. Savchynskyy A Comparative Study of Graph Matching Algorithms in Computer Vision ECCV 2022 [pdf] [project page]
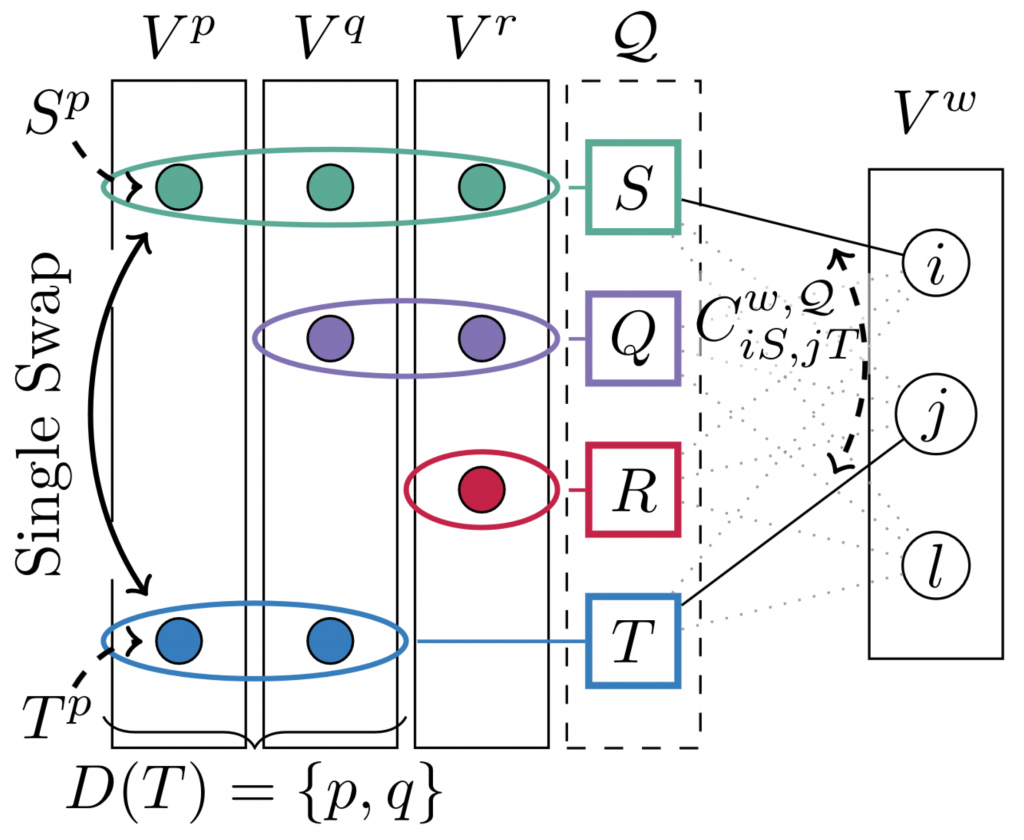
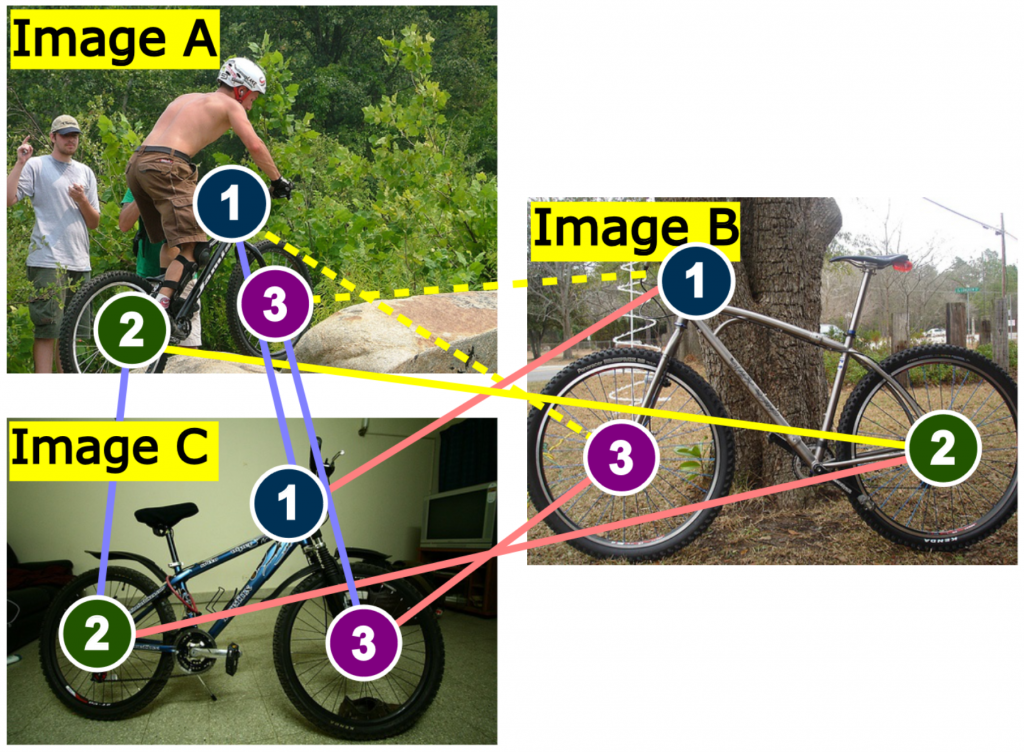
Unsupervised discrete cycle-loss-based learning for graph matching
Siddharth Tourani, Carsten Rother, Muhammad Haris Khan, Bogdan Savchynskyy Unsupervised Deep Graph Matching Based on Cycle Consistency AAAI 2024 [extended version] [code] [poster]
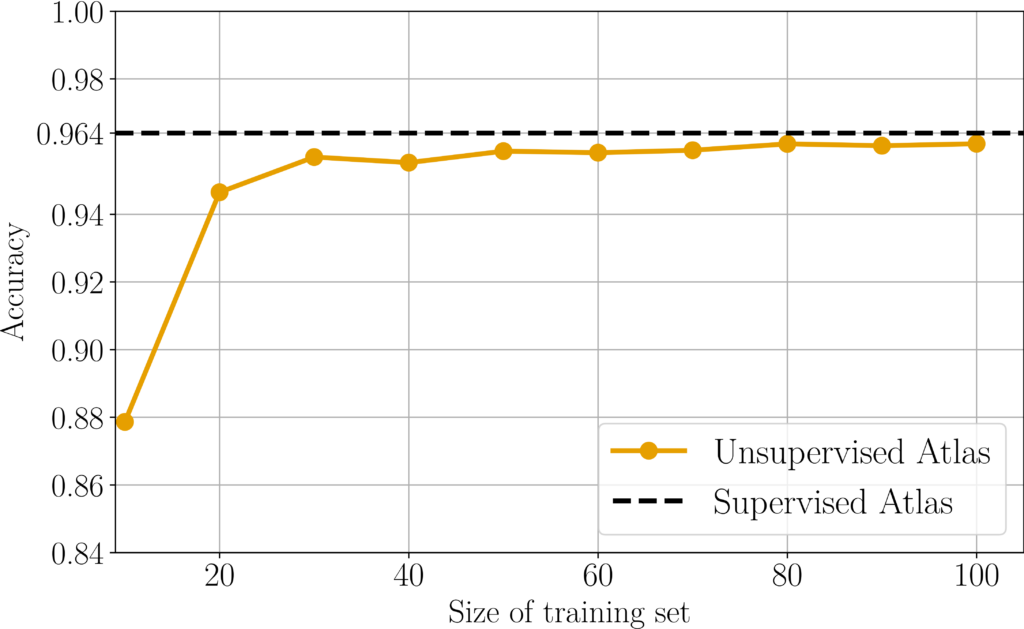
Fully unsupervised atlas of C. Elegans with nearly supervised accuracy
Christoph Karg, Sebastian Stricker, Lisa Hutschenreiter, Bogdan Savchynskyy, Dagmar Kainmueller
Fully Unsupervised Annotation of C. Elegans arXiv:2503.07348 [pdf]
